HLA Epitope Matching for Mere Mortals
Human leukocyte antigen (HLA) epitope matching is something you’ve heard about but you can’t exactly remember the specifics. You might remember it as a contender in the Transplant Basic Science region of NephMadness 2016; however, you cannot quite remember the difference between an eplet and an epitope and how exactly it fits in with traditional serological HLA subclasses. A recent review published in AJKD by Sypek et al aims to explain these concepts to the general (and transplant!) nephrologist.
The authors begin by presenting a history of the discovery of the HLA system from serological subtypes to molecular typing to high-resolution modern sequencing revealing thousands of distinct HLA alleles.
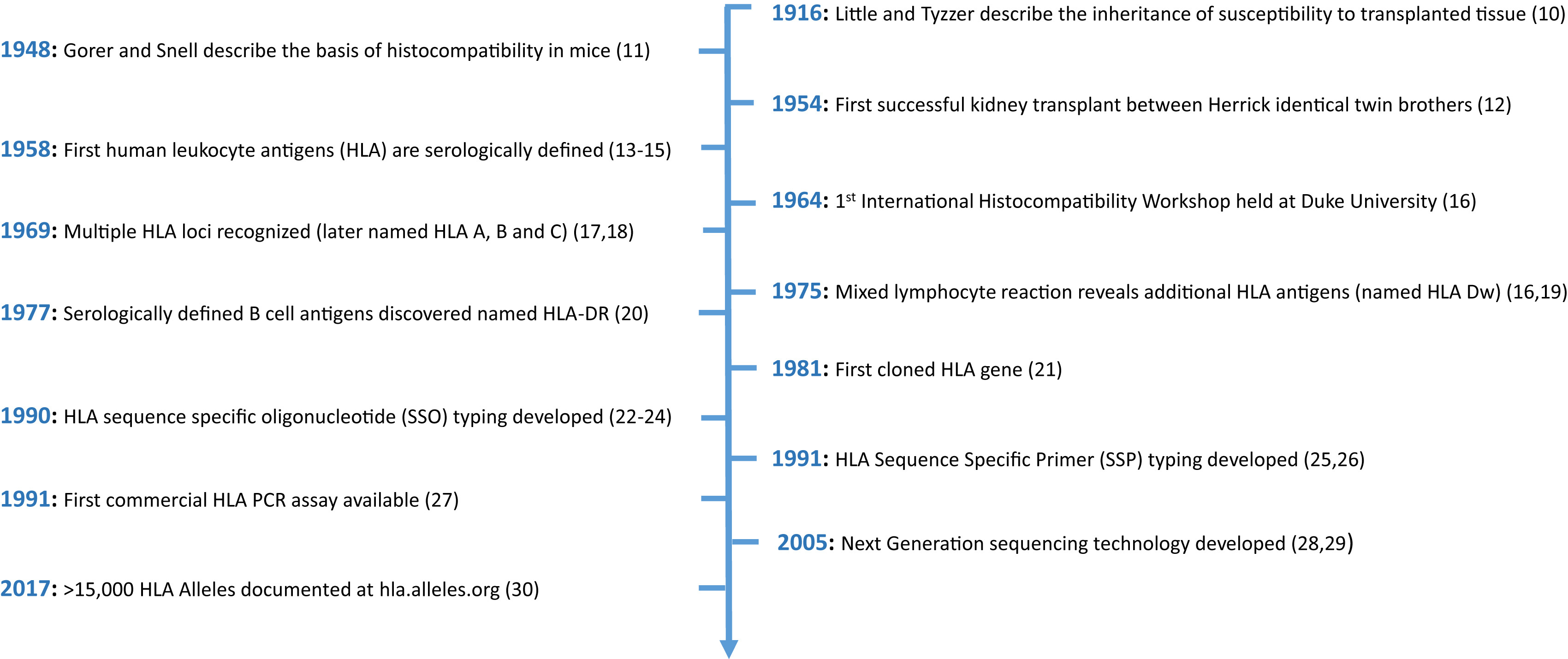
Timeline of key discoveries relating to HLAs. Figure 1 from Sypek et al, AJKD © National Kidney Foundation.
Whilst the impact of HLA mismatching on graft survival may have been abrogated by modern immunosuppression regimes, it remains a significant predictor of acute rejection and graft survival. Moreover, donor/recipient HLA mismatch leads to risk of donor-specific antibody formation, which may be deleterious to the allograft.
Clinicians have come a long way since 1958, when the first HLAs were identified when white blood cells were mixed with the serum of multiparous women and patients receiving multiple transfusions. Over time, HLA-B and HLA-C subgroups were discovered, and techniques to define HLA groups into specific subtypes and linked genetic loci (DR, DQ, and DP) were known by the mid-1970s.
With our increasing knowledge of the complexity of the HLA system, relying on low-resolution typing to make allocation or treatment decisions is unwise. For example, anti-HLA antibodies detected by modern single bead assays may not necessarily bind to the corresponding crude HLA subtype. Erroneously excluding HLA antigens from consideration, particularly for highly sensitized individuals, would adversely affect organ offering. Sypek et al also point out that the current system fails to account for the immunogenicity of a specific mismatch (the ability to induce an antibody response) or the ability for antibodies to cross react with other binding sites.
The authors describe HLA epitope matching as a much more precise reflection of actual antibody interaction with antigen. HLA antigens contain strings of polymorphic sites (epitopes) that serve as target regions for antibody binding (see image below).
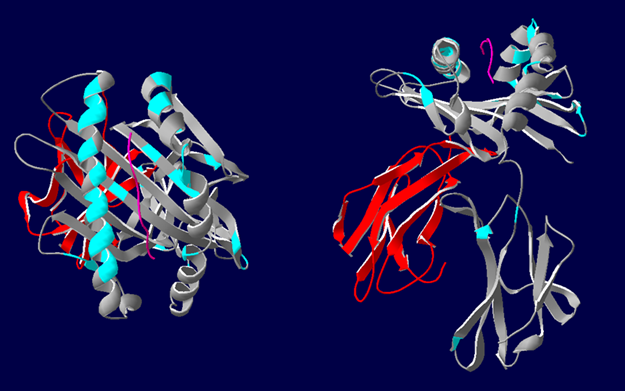
3D representations of HLA-A*02 showing the polymorphic regions accessible for antibody binding. β2-microglobulin (red), HLA class I α-chain (grey) with epitopes highlighted in blue. Most polymorphic regions are focused around the peptide (pink). Image courtesy of Dr. Richard Battle, Histocompatibility & Immunogenetics Laboratory, Royal Infirmary of Edinburgh. Created using HLA Matchmaker. From NephMadness 2016.
Rather than binding to the whole antigen, antibody binds to specific epitopes not present on self-HLA antigens. Each epitope may bind a number of distinct antibodies, explaining the cross-reactivity mentioned above. It may also explain why foreign HLA may elicit sensitization across a broad range of seemingly unrelated antigens.
Each structural epitope contains a ‘functional epitope’ of 2–5 residues, some of which are non-self, that determine the strength and specificity of antibody binding. These were initially described as linear amino acid sequences named ‘triplets.’ Subsequent analyses have included discontinuous sequences brought together by protein folding. The term “eplet” is used to describe these polymorphic residues (some of which are theoretical) which act as the functional epitopes. Defining HLA mismatch at the eplet level allows for a more detailed assessment of HLA compatibility, and can be performed using freely available software (eg HLA Matchmaker).
Despite these advances, there are still gaps in our understanding of this subject. For example, counting up the number of eplet mismatches and considering them all equal does not appear to make biological sense. We already know that determining immunogenicity of a mismatch is a complex process, with the physicochemical properties of the epitope mismatch being proposed as another method of assessing this (electrostatic mismatch score; the sum of the differences in isoelectric point for each mismatched amino acid).
Organ allocation systems are beginning to incorporate epitope matching in an effort to enhance compatibility. For example, the Eurotransplant Acceptable Mismatch program aims to prioritize appropriate kidneys for highly sensitized recipients who would otherwise have unacceptably long waiting times. Work is being done to define epitope mismatch thresholds for de-novo donor specific antibody (DSA) formation as well as allograft function. Despite the promise that epitope matching offers in decreasing sensitization and DSA formation, we have much still to learn.
The literature in renal transplantation is succinctly reviewed in this manuscript, which provides enough detail for the transplant nephrologist while still clearly explaining the concepts for the interested generalist. This is a fascinating field which will hopefully bring improvement to clinical care for our transplant patients.
– Post prepared by Paul Phelan, AJKDBlog Contributor. Follow him @paulphel.
To view Sypek et al (subscription required), please visit AJKD.org.
Title: HLA Epitope Matching in Kidney Transplantation: An Overview for the General Nephrologist
Authors: M. Sypek, J. Kausman, S. Holt, and P. Hughes
DOI: 10.1053/j.ajkd.2017.09.021

Leave a Reply