#NephMadness 2017: Biomedical Research Region

Copyright: Sebastian+Duda / Shutterstock.com
Submit your picks! | NephMadness 2017 | #NephMadness or #BioMedRegion
The 2017 edition of NephMadness features 4 techniques that have or will alter the face of biomedical research. This is a who’s who of cutting-edge research, all with great potential to revolutionize the diagnosis and treatment of kidney disease. Will it be the run and gun UNLV style Team Gene Array or will we reinvigorate the field like Coach Calipari did with multiple one and done’s with the specific and unbiased Team Single Cell RNA Sequencing? We are not done yet. The most heralded team to make the field has to be CRISPR-Cas9. Team CRISPR has the most potential to be a game changer. Will they live up the the hype and “cut” the field wide open? We will have to see. Lastly, we have Team 2-Photon. The ability to image living tissue was a huge advance and will allow for deeper understanding of physiology. Which team will reign supreme in the biomedical research region? Let’s take a look at the detailed scouting reports.
Selection Committee Member for the Biomedical Research Region:
Benjamin D. Humphreys, MD, PhD
Dr. Humphreys is the Joseph Friedman Associate Professor of Renal Diseases in Medicine and Chief, Division of Nephrology at Washington University in St. Louis. In his basic science laboratory, Dr. Humphreys investigates the mechanisms of kidney injury and repair using genetic models, pluripotent stem cells, and single cell sequencing approaches. Follow @HumphreysLab on Twitter.
Competitors for the Biomedical Research Region
Gene Microarray vs Two Photon Microscopy
CRISPR-Cas9 vs Single Cell RNA Sequencing
Gene Microarray

Copyright: science+photo / Shutterstock.com
Team Gene Microarray is like the UNLV of the 90s. Just as UNLV tore through the competition in the 80s under the helm of Jerry Tarkanian, you can’t open a paper and not see a gene microarray. Huge advances in molecular techniques allowed for the simultaneous measurement of multiple mRNAs in this platform. Although techniques like RNA sequencing have started to encroach on its territory, gene microarray is still a workhorse of biomedical research. These highly parallelized assays are useful to identify differences in gene expression: for instance, differential gene expression can be characterized in the podocytes of a mouse model of FSGS (Actn4-/-) and compared to wild-type control in order to find novel therapeutic targets to exploit. Another example is a microarray performed in diabetic human glomeruli and healthy controls (see figure) which revealed a number of dysregulated genes. So, what are the nuts and bolts of gene microarray?
The technique:
- Tissue extraction, purification of RNA using a variety of techniques
- Analysis of purity and quantification
- Reverse transcription of mRNA into complementary DNA (cDNA)
- Hybridization of cDNA (tagged with a fluorescent molecule) to pre-designed DNA probes that are patterned on an array
- Scanning of the microarray and quantification of the signal
- Total amount of signal is related to the abundance of gene products or mRNA
- Comparison of signals between controls and experimental groups (as shown in figure below)

Fig 3 from Baelde et al, AJKD, © National Kidney Foundation. A dendrogram of unsupervised hierarchical clustering on the basis of similarity in gene expression patterns of the 6 different arrays show the degree of relationships of samples. Different colors show the normalized z score for each gene. Green indicates upregulated genes, and red indicates downregulated genes. Abbreviations: C1a, control 1; C1b, duplicate of control 1; C2, control 2; D1a, diabetes 1; D1b, duplicate of diabetes 1; D2, diabetes 2.
Thus, microarrays are capable of looking at the relative expression of thousands of genes. The Renal Gene Expression Database (RGED) has been generated of gene expression profiles in kidney disease, and researchers in Innsbruck, Austria provide a wealth of data on functional genomics and biomarker research in kidney disease. While, the number of studies using microarray in their research is mind boggling, the main issue with microarray is the need to prespecify the genes you are assaying. This is in direct contrast to RNA sequencing which is an unbiased approach and will theoretically give you data on all RNAs transcribed. Affymetrix is a popular maker of microarrays and these can be customized for your needs. For instance you can order an assay for just the genes are known to be associated with podocyte health and disease. There’s no question that in the NephMadness field, microarray is still a formidable challenger. With a mature roster stocked with seniors we know to expect with this team.
Two Photon Microscopy

Copyright: WH+CHOW / Shutterstock.com
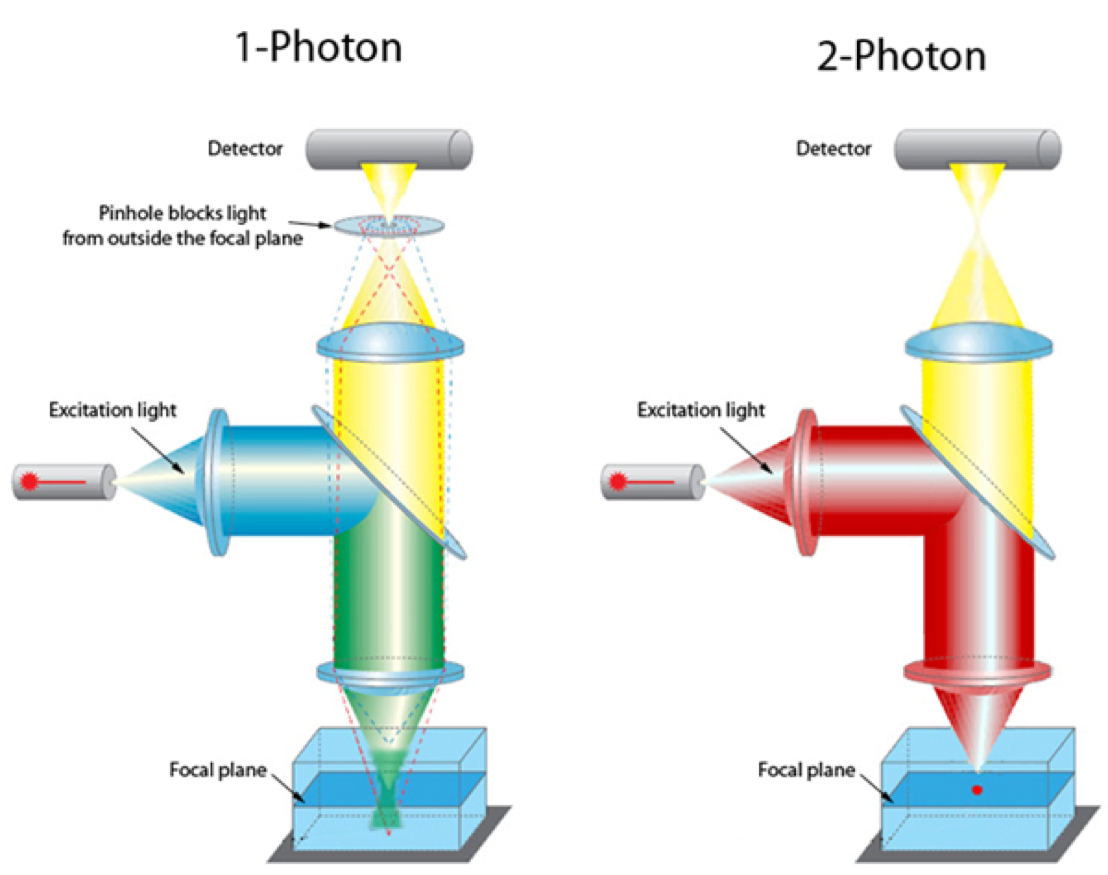
Imagine looking into a functioning kidney and seeing glomerular filtration, blood flow, cell-cell interactions, and subcellular processes highlighted by fluorescent tracer molecules and biosensors. The use of sophisticated microscopy to allow imaging of intact organs rather than just cells grown in culture is revolutionizing biomedical research. The next important advance has been a technique to image living tissue without the phototoxicity that accompanies standard confocal imaging. What is it that makes 2-photon microscopy able to do this? Let’s look at 2 key advantages of 2-photon vs 1-photon confocal microscopy.
- First, the use of longer wavelengths and the requirement that two lower energy photons must strike the target allows deeper penetration into tissue and no out-of-focus fluorescent excitation and emission thus reducing phototoxicity.
- Second, the 4-dimensional (time) nature of the image obtained allow for dynamic imaging of physiologic and pathologic processes.
Before we get too technical, let’s take a moment and go back in time. The concept was first described by Maria Goeppert-Mayer during her doctoral dissertation in 1931. She was the second woman to win a Nobel prize (first was Marie Curie, who collected two, so though Maria was the second woman, she received the third Nobel prize awarded to a woman).
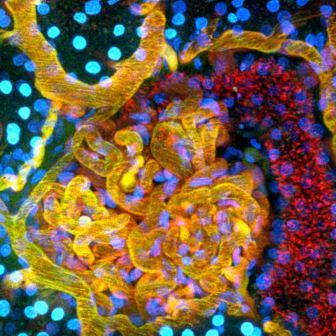
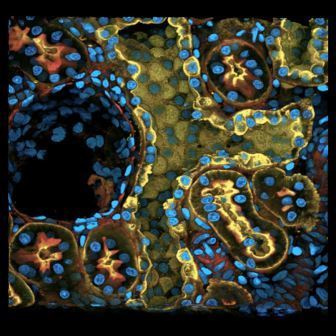
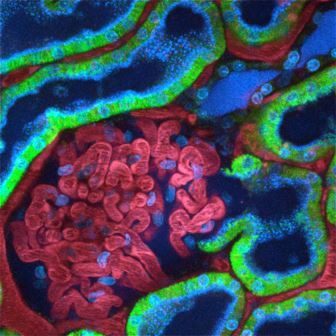
Let’s compare and contrast 2-photon with confocal microscopy. Conventional confocal requires a single high-energy photon (shorter wavelength) to deliver enough energy to induce the excited state, and when it returns to the ground state it emits a photon of fluorescence. Thus, excitation occurs above and below the plane of focus resulting in a blurred image. In order to focus the image an adjustable pinhole is used to reject out-of-focus fluorescence (see figure). In contrast, 2-photon microscopy requires 2 lower-energy wavelength photons (longer wavelength) to strike the fluorophore simultaneously in order to induce the excited state and cause emission of a photon. In essence this is a kind of optical sectioning as there is only one focal plane where there are enough focused photons to result in fluorophore activation. Thus, 2-photon is particularly attractive for intra-vital (living) fluorescence imaging. The use of 2-photon microscopy has provided breathtaking microscopic views of the kidney, allowing a finer understanding of the relationship between kidney struction and function.
The Indiana O’Brien Center for Advanced Microscopic Analysis is funded by a NIH P30 O’Brien center grant. Images and videos for instructional purposes are available for downloading from their website and a workshop for interested investigators is forthcoming. This disruptive technology has made important quantitative contributions to the assessment of many physiologic and pathologic kidney processes (see website).This degree of dynamic insight into living tissue compartments and structure-function relationships is unprecedented. 2-photon is likely to “see” a long run in NephMadness 2017.
- Collecting Duct image via Indiana O’Brien Center for Advanced Microscopic Analysis / Indiana University
- Glomerulus image via Indiana O’Brien Center for Advanced Microscopic Analysis / Indiana University
- Glomerulus image via Indiana O’Brien Center for Advanced Microscopic Analysis / Indiana University
CRISPR-Cas9

Copyright: GeK / Shutterstock.com
NephMadness 2017 is filled with teams steeped in history, legacy, and drama. With CRISPR comes something we haven’t seen before. The emergence of a game changer in gene editing. Many kidney diseases are known to be attributed to mutations in specific single genes (ADPKD, Fabry, FSGS, Liddle, etc). Can CRISPR be harnessed to fix these mutations? How can this technology be used in the research lab to advance our understanding of physiology and more specifically kidney diseases? What exactly is CRISPR-Cas9 anyway, and didn’t we already have a way to do this? Why is this different and is the hype justified? Does this team of diaper dandies have what it takes to reach one shining moment? Let’s find out.

Well, let’s begin with some basics. CRISPR-Cas9 is an elegant bacterial system that confers immunity to foreign pathogens. Before we discuss how it has been harnessed in biomedical research, we need to see how this system works in bacteria. Basically, this is their immune system and is constantly scanning the bacterial genome for foreign DNA and then excising it. The system has two components. First, is the CRISPR, which is an acronym for
- clustered
- regularly
- interspaced
- short
- palindromic
- repeats
These are segments of DNA in the bacterial genome that allow for the steady production of short sequences of foreign viral or plasmid DNA. These short segments complex with the enzymatic scissor known as Cas9, or CRISPR associated protein 9. It’s essentially a nuclease that cuts DNA and RNA at sites to which it is instructed to bind. How does it know where to cut? It is guided by “guide RNAs”, or gRNAs, that are complementary to specific sites in the genome. In the case of bacteria, the gRNAs direct Cas9 to foreign DNA sequences. By constantly scanning the host genome for foreign DNA, and then directing the Cas9 nuclease to foreign DNA insertions via gRNAs, CRISPR-Cas9 detects and removes virus and plasmid invaders in the bacterial genome.
Researchers took these tools found in bacteria and applied them to eukaryotic cells in the laboratory as a way to manipulate DNA. All of the components are there. Another shining example of how understanding of basic mechanisms (this time in bacteria) can have far-reaching effects. If you are interested in targeting a gene all you need to do is make the short guide RNA that is specific to the sequence you are targeting (and not to other sequences). It is important to understand that until now, the only way researchers had to target a specific site in the genome was by homologous recombination (a Nobel prize-winning discovery, and we predict that CRISPR/Cas9 will also one day win the Nobel). Homologous recombination is HARD. It requires about 7,000 bp of DNA to target one site in the genome. These plasmids are difficult to construct and might take one researcher in the lab 6 months. By contrast, a gRNA is only 20 bp but this is sufficient to achieve the same genome specificity. This is just a brief list of potential options for CRISPR-Cas9 technology:
- Permanent knock out of a single gene
- Permanent knock out of multiple genes
- Temporary knock out of a gene
- By having mutated the Cas9 protein to lack the scissor (nuclease-null)
- Correction of a single gene mutation
- Correction of multiple gene mutations
- Assessing epigenetic mechanisms
- By fusing human acetyltransferase p300 with a nuclease-null Cas9
- Generation of chromosomal rearrangements
There are other methods of gene targeting – Cre-loxP, ZFNs, and TALENs – but the advantage of CRISPR-Cas9 is that it’s cheaper, quicker, and has higher efficiency. It is clear CRISPR-Cas9 will have a huge impact in medicine and biomedical research. The huge upside is the manipulation of genes for use in cell culture, creation of mutant animals, and eventually use in humans.
What are the downsides? The biggest downside is that CRISPR-Cas9 can have off-target effects. The targeting gRNA can potentially recognize similar but not identical sequences and introduce mutations. These can be difficult to identify but efforts to scan regions of the genome with some similarity to the gRNA are typically performed to ensure this doesn’t happen. There is also the potential for mosaicism if the gene manipulation occurs beyond the one-cell stage of embryonic development. If multiple embryos are targeted at the same time you can have offspring with different gene manipulations since the DNA repair mechanism randomly inserts base pairs into the cut DNA.
Very recently, a National Academy of Sciences panel made an astounding pronouncement – that ‘gene editing’ with CRISPR-Cas9 might be ethically acceptable in human embryos in certain limited situations.
What are some examples of CRISPR-cas9 being utilized in nephrology? First, CRISPR-cas9 is used to manipulate cell lines in many laboratories. Freedman et al utilized this technique to directly differentiate human pluripotent stem cells to kidney organoids. In this same study the group generated biallelic knock out of PKD1 and PKD2 in these kidney organoids and observed development of cyst-like structures. This was an important paper because it demonstrated that this approach could be used to model kidney disease in an in vitro model. Pereira et al utilized CRISPR-Cas9 to introduce alpha-galactosidase A deficiency into a human podocyte cell line to model Fabry disease.
The use of CRISPR-Cas9 is like UCLA freshman Lonzo Ball, who’s just 18 but one of the most impactful players in the game and has the potential to rock the NCAA and NBA. CRISPR-Cas9 will have huge impact in the further understanding of kidney disease. The long-term implications as this system is further refined is that gene mutations could eventually be targeted to the culprit cell. Human clinical trials using CRISPR-Cas9 are already underway. Does CRISPR have what it takes to make a long run in NephMadness 2017? We will all be watching this one closely.
Single Cell RNA Sequencing

Copyright: ktsdesign / Shutterstock.com
Another freshman biomedical tool that is making waves is single cell RNA sequencing. What the heck is that? This is a powerful way to look at differences between individual cells to understand which genes are activated and which are downregulated. Until recently, using techniques like microarray, scientists could only examine RNAs in bulk populations of cells – in fact, entire tissues comprising dozens of different cells. Which cell expresses which RNA? It was hard to tell.
The power in this very new approach is that we can now understand – at the level of an individual cell – conceivably all of the changes that govern cell identity. The old method was all about taking a piece of tissue with hundreds of different types of cells and assaying differences in “known” gene transcription with probes. Like the rejuvenated Kentucky under the helm of John Calipari, single cell RNA sequencing gets all of the new recruits. Let’s review this technique and discuss what makes it so powerful…
Before we get the RNA sequence we must first explore the advances in single cell isolation. The most notable and widely used technique is flow-activated cell sorting (FACS). FACS using fluorescently labeled antibodies to isolate cells of interest according to a specific cell-surface marker. Up to 17 different antibodies can be used at one time. However, this technique is limited by the need for single cell suspensions and the requirement of specific antibodies to characterize cells. Other techniques are listed below:
- Micromanipulation– a glass micropipette to aspirate single cells under a microscope
- Optical tweezers– a highly focused laser beam to hold and manipulate
- Laser capture microdissection- technique used to isolate certain cells under a microscope
Let’s talk about RNA sequencing and why this is such a powerful technique. The overarching goal is to profile the type and abundance of mRNA transcripts that are being made at a given time. This has been termed the transcriptome and is an unbiased approach to screening for mRNA transcripts since you do not need to rely on known probes like with gene microarray. This unbiased approach can be extremely useful to give important clues as to how gene programs are behaving either in a physiologic or pathophysiologic manner. Older techniques, quantitative/qualitative reverse transcriptase polymerase chain reaction (qRT-PCR) or gene microarray, rely on the multiplexing with known probes (sequences). The problem there is that you are limited to the numbers of genes you can assay. RNA sequencing makes complementary DNA for all RNA transcripts that are made in the cell at a given time. This includes both coding and non-coding RNA (these are typically never assayed by other methods). Here is how the technique works:
- RNA is reverse transcribed to make first-strand cDNA
- second-strand synthesis is performed and the cDNA amplified
- cDNA sequencing is performed using next-generation sequencing technologies
- This is repeated on hundreds to thousands of cells
- Big data processing
The NIDDK recognized the potential for this approach and launched the NIDDK Kidney Precision Medicine Project (KPMP). The goals of this initiative are to:
- Evaluate human kidney biopsies from participants with AKI or CKD
- Create a kidney tissue atlas
- Define disease subgroups
- Identify critical cells, pathways, and targets for novel therapies
NIDDK thinks that if we can deconvolute the single cell transcriptomes from the full spectrum of kidney disease (CKD, AKI, FSGS, IgA, etc) that we will better be able to provide patients with personalized prognosis, and therapeutic options. Therefore, single cell RNA-sequencing is perfectly aligned to unlock the answers to these important questions and provide novel therapeutics. Applications are already in and awards should be announced soon. This technique is so new that there are few if any single cell transcriptome papers in nephrology yet. So consider how lucky you feel, and what impact this might have, in voting for this hot new technology!
– Post written and edited by Matt Sparks (@Nephro_Sparks).





Leave a Reply